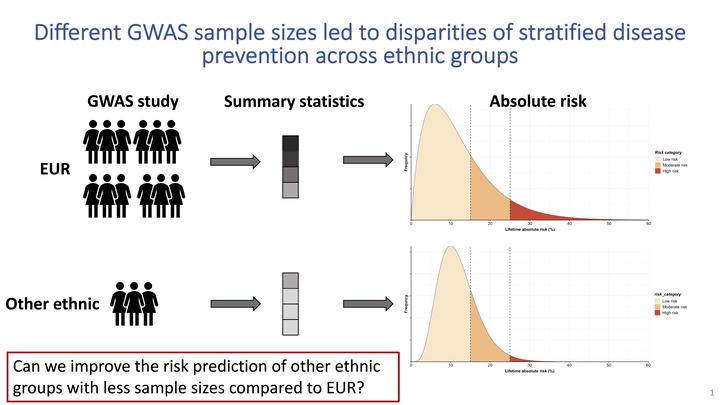
Polygenic risk scores (PRS) are useful for predicting various phenotypes/outcomes; however, most PRS are developed using predominately European ancestry data, their performance in non-European populations is often poorer. To improve PRS performance in non-European populations, we propose TDLD-SLEB (Two-Dimensional Clumping and thresholDing with Super Learning and Empirical Bayes), which takes advantage of both existing large GWAS from European populations and smaller GWAS from non-European populations. TDLD-SLEB leverages the genetic correlations across populations while accounting for the linkage disequilibrium (LD) differences and population-specific allele frequencies. We found TDLD-SLEB had outperformed alternative methods through large scale simulations with different genetic architectures. Using 23andMe, Inc. data, we applied TDLD-SLEB for seven complex traits using GWAS data from five ethnic groups (average N≈ 3,108K per trait). TDLD-SLEB often led to large improvement in PRS performance compared to alternative methods for African American population (e.g., for height, R2 = 0.12 for TDLD-SLEB vs. R2 = 0.05 for weighted PRS method). For other ethnic groups, TDLD-SLEB also led to sometime notable improvements in PRS performance, such as for cardiovascular disease in the Latino population (AUC = 0.61 for TDLD-SLEB vs. AUC = 0.58 for weighed PRS method). In conclusion, TDLD-SLEB is a computationally scalable and statistically efficient method for generating predictive PRS in non-European populations.